Which form of dengue evolved first?
We know that the dengue virus comes from mosquitoes and has four different forms but which of its four forms arose first? This question is answered below, intentionally in a long form to illustrate basic concepts in Bioinformatics and how python can be effectively used for different types of analysis in Bioinformatics.
The impact of dengue
Dengue fever is a mosquito borne disease caused by the dengue virus (DENV). It occurs in about 100 countries in the world and about 3 Billion people (almost half the world's population!) is at risk. A 2013 paper in Nature estimates that DENV infects about 390 Million people in the world every year, of which about 96 Million manifest apparently any level of clinical or sub-clinical severity. The mortality estimate is 1-5% without treatment and less than 1% with adequate treatment. However, the more severe form of dengue fever called the dengue hemorrhagic fever which occurs in less than 5% of all dengue cases , carries a mortality rate of about 26%. The virus that causes dengue disease is divided into four closely related serotypes (dengue virus 1, 2, 3 and 4). Researchers have shown that the more dangeous hemorrhagic fever occurs when a person, after being infected with one serotype, gets infected with another one. The wikipedia picture below shows a TEM micrograph with dengue virus virions (the cluster of dark dots near the center, each about 50nm in size). Below that the structure of the outer shell of a single DENV virus is shown (from PDB). 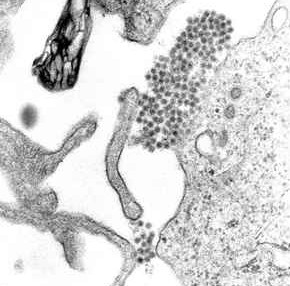

Flaviviruses
After the scary and alarming description above, now we come to the main question: Where did the Dengue virus come from? To answer this question, we will go through some basics. Dengue viruses belong to a virus family called Flaviviridae which includes, among other viruses, the West Nile and Hepatitis viruses. To further the impact this virus family has on humans, note that about 2-3% of the world population (this number is equal to Pakistan's current population!) is infected with Hepatitis C. No vaccine exists either for Dengue or Hepatitis C and this presents a huge risk.
Biologically, these viruses are RNA viruses, i.e., they use RNA as their genetic material instead of DNA (as in humans). You can thus think of the genetic material of these viruses as a 4 character alphabet (the 4 nucleotide bases A,C,G and U) string with length of around 11,000. Theirs is a really small genome in comparison to, say, humans (3.2 Billion DNA base pairs) and the canopy plant (150 Billion DNA base pairs). The genomic sequence of an organism can be found through genomic sequencing technologies. Off the topic: It is interesting to note that each of the 100 trillion or so cells that humans have contains a dust particle sized cell nucleus (typical diameter of 2-10 microns) a small part of which (chromosomes) stores 3.2 Billion base pairs of pragmatically and simultaneously accessible genetic information. Now, that is some serious information density!
RNA and proteins
Biological cells and viruses function by using the RNA as a pseudo-template to generate proteins through the conceptually straight-forward process of translation. Proteins are large molecules made up of long linear chain(s) of amino acid residues which folds into a 3D structure. With a tilt of the hat to DNA and RNA, proteins are the most important biological molecule as all vital cellular or viral processes ultimately involve proteins. For example, the translation of RNA into proteins is itself made possible through proteins. The outer shell of the virus is also made up a mosaic of a complex of three proteins. In flaviviridae, the replication of the virus involves two proteins called NS3 and NS5. To appreciate the importance of proteins in general, note that 50% of the dry weight of the human body is protein. As a computer scientist, you can think of a protein as an arbitrary length string of 20 characters (the 20 amino acids). For example, the NS3 protein in DENV is 619 amino acids long.
Since all proteins are coded through RNA, so theretically, the protein sequence contains the same information as the coresponding RNA sequence. As a matter of fact, the sequence of all proteins in a virus (called the polyprotein) is often used instead of the genome.
The proteins of dengue
Below, we first display, using Python's Biopython package, the polyprotein sequence of DENV downloaded from uniprot as a FASTA file.
from Bio import SeqIO
import urllib2 as url
def downloadFile(url,ofname=None):
import urllib2
if ofname is None:
ofname = url.split('/')[-1]
u = urllib2.urlopen(url)
with open(ofname, 'wb') as f:
meta = u.info()
file_size = int(meta.getheaders("Content-Length")[0])
print "Downloading: %s Bytes: %s" % (ofname, file_size)
file_size_dl = 0
block_sz = 8192
while True:
buffer = u.read(block_sz)
if not buffer:
break
file_size_dl += len(buffer)
f.write(buffer)
status = r"%10d [%3.2f%%]" % (file_size_dl, file_size_dl * 100. / file_size)
status = status + chr(8)*(len(status)+1)
print status
return ofname
def readFASTA(fname):
"""
Reads the fasta file fname and returns the sequence object
"""
with open(fname, "rU") as handle:
record = list(SeqIO.parse(handle, "fasta"))
return record
url = 'http://www.uniprot.org/uniprot/P29990.fasta'
records = readFASTA(downloadFile(url))
print "\nThere are",len(records),"records in this file\n"
# Read the first one
r = records[0]
print "The length of dengue polyprotein is: %i \n" %len(r)
print "The name of the record is:",r.name,"\n"
print "The description of the record is:",r.description,"\n"
print "The polyprotein sequence is shown below:\n"
print str(r.seq)
The NS3 proteins of different flaviviruses
We can also view the protein sequnces of NS3 proteins from different flaviviruses stored in the file NS3.fasta.
fname='NS3.fasta'
records = readFASTA(fname)
for r in records:
print r.id, 'length is', len(r)
print
print str(r.seq)
print
As you can see, the NS3 protein from different flaviviruses are of different lengths and are different in their sequence. The NS3 proteins from different dengue virus serotypes are shown below:
dengue_records = [r for r in records if 'dengue' in r.id.lower()]
for r in dengue_records:
print r.id, 'length is', len(r)
print
print str(r.seq)
print
Sequence alignment: Measuring similarity between protein sequences
The answer to the question of which dengue came first, lies in exactly how similar these proteins (or overall virus polyproteins) are to eachother. Once we know the complete genomic sequence or the sequence of amino acids of one or more proteins, we can use this information to find out how similar one organism is to another. This is done through the bioinformatics technique of sequence alignment. Sequence alignment algorithms like Smith-Waterman, Needleman-Wunsch or BLAST, given two sequences, gives you a scor of how similar the two sequences are and how they align with respect to each other. These algorithms return a score which is indicative of how good the alignment is. Below, we align, using Biopython, the NS3 protein sequence of a dengue virus serotype against the corresponding proteins from West Nile and Hepatitis C virus.
import Bio.pairwise2 as pairwise2
from Bio.SubsMat import MatrixInfo as matlist
from time import time
def algin(s1,s2):
matrix = matlist.blosum62
gap_open = -10
gap_extend = -0.1
start = time()
alns = pairwise2.align.localds(s1, s2, matrix, gap_open, gap_extend)
aln = alns[0]
print "alignment done in %.2f seconds" %(time()-start)
aln_s1, aln_s2, score, begin, end = aln
print 'sequence 1:',s1.id,':',len(s1),'\n'+str(aln_s1.seq)
print 'sequence 2:',s2.id,':',len(s2),'\n'+str(aln_s2.seq)
print 'alignment score:',score
denv = dengue_records[0]
wnv = [r for r in records if 'west_nile' in r.id.lower()][0]
hepc = [r for r in records if 'hepatitis' in r.id.lower()][0]
print '*'*20
print 'DENV-WNV'
algin(denv,wnv)
print '*'*20
print 'DENV-HEPC'
algin(denv,hepc)
As you can see above, the DENV NS3 is more similar to the WNV NS3 than the HEPC NS3.
Phylogenetic trees
We can compare all the virus species using the same way and then, based on the similarities, construct a tree (formally caleld a phylogenetic tree) which shows how each viral species relates to the other: two viruses that are similar should be grouped into the same branch of the tree. Generation of this tree requires multiple sequence (or all pair) pairwise alignments.
The alignment algorithm used above is a deterministic Dynamic programming algorithm and is, consequently, quite slow. Even though Biopython supports the generation and display of phylogenetic trees, we use the much faster MEGA6 package which can not only generate multiple sequence alignments using CLUSTALW but can also generate phylogenetic trees.
The code below, uses the NS3.fasta file to generate a multiple sequence alignments in its own '.meg' format and a tree in the newick file format. It demonstrates how easy it is to use Python as glue.
import os
def getFileParts(fname):
"""
Returns the parts of a file (path,name,ext)
"""
(path, name) = os.path.split(fname)
n=os.path.splitext(name)[0]
ext=os.path.splitext(name)[1]
return (path,n,ext)
class Mega:
"""
Wrapper for the Mega 6 Computational Core program for Phylogenetics
You can download the Mega package from: http://www.megasoftware.net/megacc.php
Use the Mega 6 analysis settings prototyper (included in the mega package) to generate the settings files
"""
def __init__(self,megaexe='M6CC.exe'):
"""
megaexe is the path to the executable
"""
assert os.path.isfile(megaexe), "Mega executable missing" #file must exist
self.mexe = megaexe
def align(self,settings,ifile,ofile=None):
"""
Generation of alignments using Mega
input:
settings: path to settings file for alignment
ifile: path to input fasta or sequence file
ofile: path to output file (if not specified, name automatically generated)
output:
result: whether the exe was run successfully (0) or not
ofile: path to the output .meg file
"""
if ofile is None:
opath,oname,oext = getFileParts(ifile)
ofile = os.path.join(opath,oname+'_aln')
if os.path.isfile(ofile+'.meg'):
print 'Warning:', 'existing file',ofile+'.meg','will be removed.'
os.remove(ofile+'.meg')
cmd = self.mexe + ' -a '+settings+' -d '+ifile+' -o '+ofile
result = os.system(cmd)
ofile+='.meg'
return result,ofile
def phylo(self,settings,ifile,ofile=None):
"""
Generation of phylogenetic tree using Mega
input:
settings: path to settings file for tree generation
ifile: path to input multiple sequence alignment (.meg) file
ofile: path to output newick file (if not specified, name automatically generated)
output:
result: whether the exe was run successfully (0) or not
ofile: path to the output .nwk file
"""
if ofile is None:
opath,oname,oext = getFileParts(ifile)
ofile = os.path.join(opath,oname+'_phy')
if os.path.isfile(ofile+'.nwk'):
print 'Warning:', 'existing file',ofile+'.nwk','will be removed.'
os.remove(ofile+'.nwk')
cmd = self.mexe + ' -a '+settings+' -d '+ifile+' -o '+ofile
result = os.system(cmd)
ofile+='.nwk'
return result,ofile
mega = Mega()
aresult,afile = mega.align(settings = 'aln_clustal_blosum.mao',ifile='NS3.fasta') #settings: using clustal default with blosum matrix
presult,tfile = mega.phylo(settings = 'tree_ml.mao',ifile=afile) #settings: using Maximum Likelihood generator of phylo tree
print 'Output alignment file generated:',afile
print 'Output phylogenetic tree file generated:',tfile
Once the files have been generated, we can use Biopython to read those files and display the tree.
from Bio import AlignIO
from Bio import Phylo
import matplotlib.pyplot as plt
def meg2fasta(ifile,ofile = None):
"""
Coverts mega format files to fasta files
"""
if ofile is None:
path,name,ext = getFileParts(ifile)
ofile = os.path.join(path,name+'.fasta')
with open(ifile,'r') as ifx, open(ofile,'w') as ofx:
for i in ifx:
if not i.strip() or '#mega' in i or i[0]=='!':
continue
if i[0]=='#':
ofx.write('>'+i[1:])
else:
ofx.write(i)
return ofile
align = AlignIO.read(meg2fasta(afile), "fasta")
tree = Phylo.read(tfile, "newick")
print align #print the alignment
tree.ladderize() # Flip branches so deeper clades are displayed at top
Phylo.draw(tree,do_show=False)
ax = plt.gca().axis()
plt.axis((0.9,7)+ax[2:])
plt.show()
Discussion
The y-axis of the plot showing the phylogenetic tree is meaningless but the x-axis is derived from the degrees of similarities between the NS3 protein from different viruses and can be interpreted as a measure of number of genetic changes or mutations in this protein over time. A split in the tree shows when two known viruses species diverged through evolution. This indicates that, out of the 4 dengue species, Dengue-4 arose before other versions of the same virus.
This plot shows how different viruses are related to one another. Even though we haven't used information about the vectors of these viruses in our analysis, the viruses are (almost) neatly clustered based on their vectors: Mosquito (Culex or Aedes) borne viruses are clustered separately from the tick borne viruses which form their own cluster. Note that the mosquito viruses (in Aedes or Culex) arose independently of flaviviruses in ticks, bats and rodents (the split at branch length ~3). It also shows that the Hepatitis virus arose independently of other types of flaviviruses considered here.
This is a simple sample of what Bioinformatics is all about -- considering biological systems as information process and using computers to solve biological problems. It also shows how python can be used for Bioinformatics.
For more details on the phylogenetic analysis of Flaviviruses, see this paper.
Similar phylogenetic analysis at a larger scale has shwon that all life is linked into a tree of life. You can get an interactive version of the tree of life here.